Ailurus vec™ (A. vec™) rethinks the vector not as a single, passive vehicle for a foreign nucleic acid sequence, but as autonomous carriers that enable autonomous screening and selecting over massive libraries of DNA codes. It is an engine to scale and accelerate biology in a tube.

The quantity and quality of data determine the limits of your ambition. Previously, gathering enough data to even just fine-tune a model took weeks. And the cost per experiment, from subcloning to expression validation, can be $$ hundreds of dollars.
Applying A. vec with next-generation sequencing (NGS), now you can easily acquire massive datasets to train predictive model from scratch, at a cost of less than $5 per wet-lab data point, in a single experiment.
This enables you to establish a robust AI+Bio flywheel internally, transitioning from tedious trial-and-error methods to a more systematic and scalable approach.
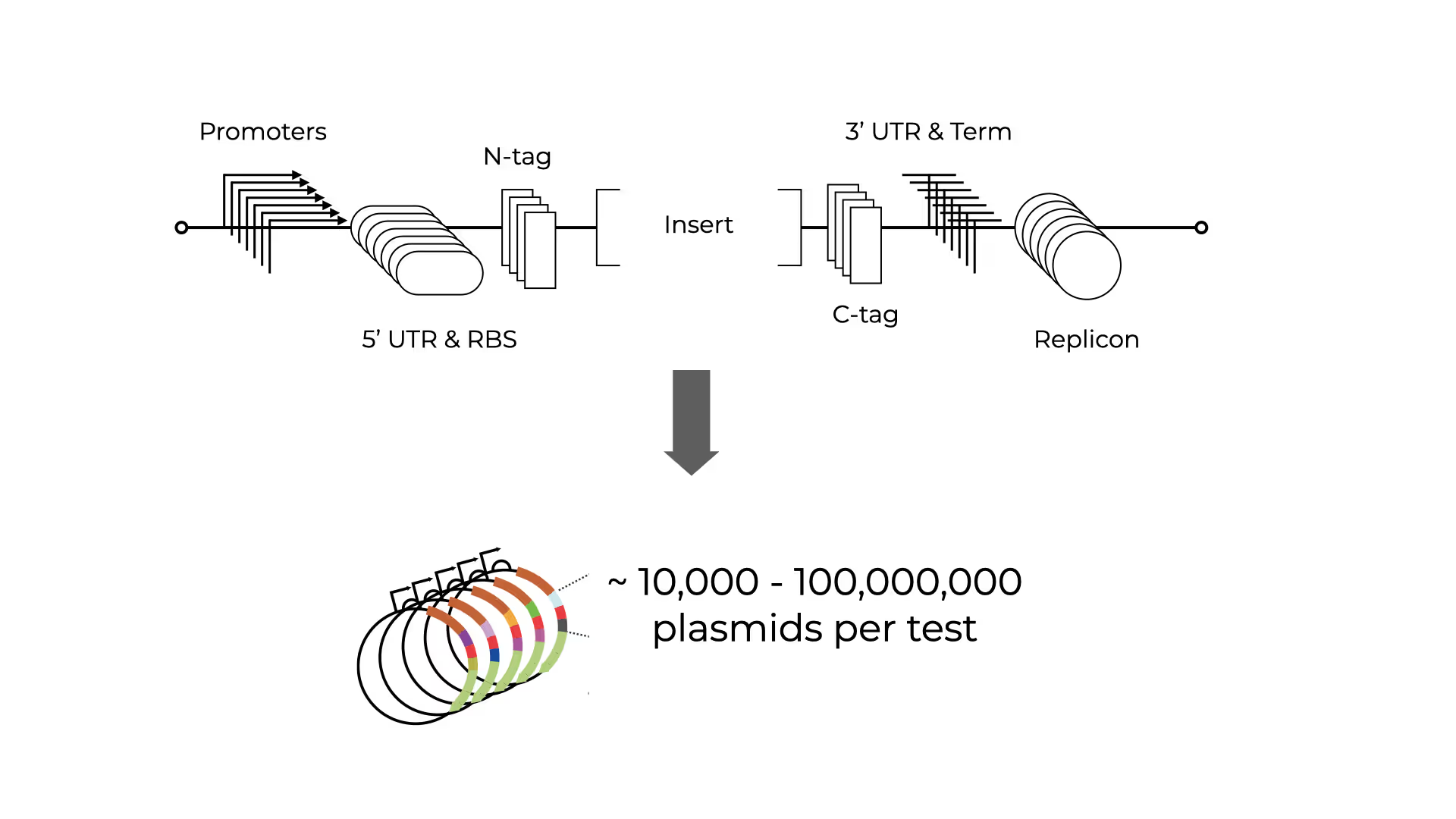
Gene expression presents a combinatorial optimization problem. A. vec E. coli Panels are assembled from pooled libraries of validated promoters, RBS, tags, terminators, and replicons, providing an extensive initial space. Our libraries vary from 10 thousand (10K Panel) to over ten trillion (10T Panel).

Biology offers a powerful solution by parallelism and selection. Each vector in A.vec panels uses a unified principle, linking target gene expression to a selection marker that only acts in its host cell. Under pressure, the culture enriches cells with the optimal vector.
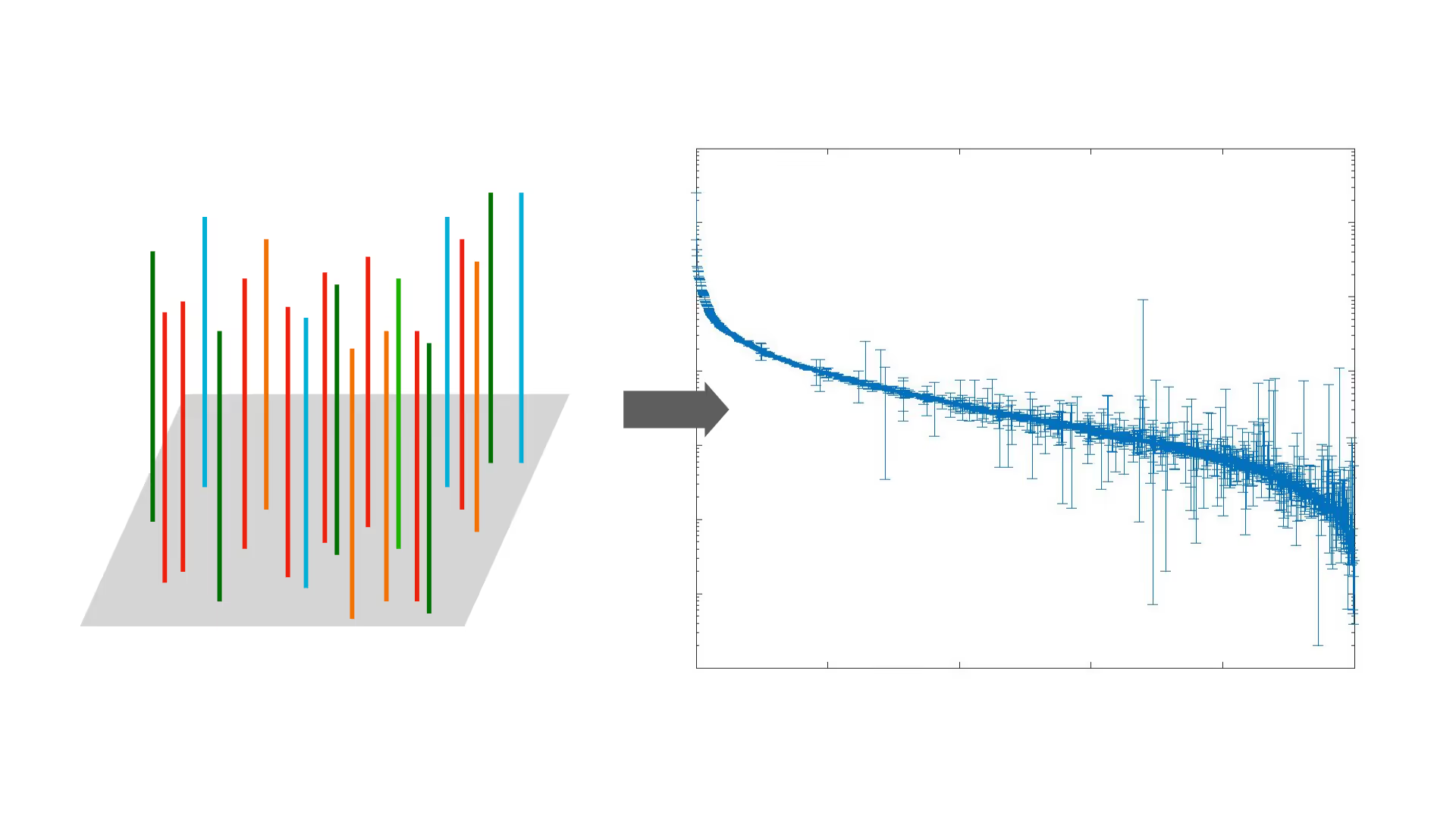
By next-generation sequencing (NGS), or other techniques (available soon), we can quantitatively measure the relative fitness of each vector in the population, which can be used for supervised learning or other AI/ML methods.

Register to the waitlist of upcoming products of Ailurus vec